Polyphosphates (polyP) are linear chains of inorganic phosphate residues linked by high-energy phosphoanhydride bonds, similar to those found in ATP. These chains can vary, ranging from a few phosphate units to several hundred.
PolyP are found in all forms of life, from bacteria and yeast to plants and animals.
In cells, polyP are involved in regulating enzyme activities, protein function, and signaling pathways.
Despite their fundamental importance, many aspects of polyP metabolism, particularly in higher organisms, remain poorly understood, making them an attractive subject of ongoing research.
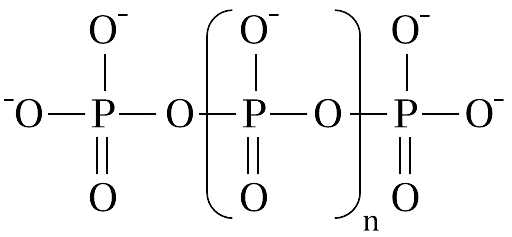
A polyphosphate structure
Protein Modification: From Monophosphate to Polyphosphate
Protein phosphorylation, the addition of a single phosphate group to a protein, is one of the most common and well-studied post-translational modifications (PTMs). However, a novel and emerging modification, protein polyphosphate modification (PPM), has recently been identified. This modification involves the attachment of polyphosphate (polyP), an inorganic polymer present across all life forms. While polyP synthesis is well-characterized in bacteria and yeast, its origin and regulation in higher eukaryotes remain largely unexplored.
Our research has uncovered a new interaction where proteins with histidine repeats exhibit a strong affinity for polyP, leading to a modification we term histidine-polyphosphate modification (HPM). This interaction is non-covalent, yet it significantly alters protein activity, hinting at its potential regulatory roles. Additionally, we have identified lysine-polyphosphate modification (KPM), which is similarly driven by ionic interactions within lysine-rich clusters.
We hypothesize that these PPMs, representing a novel class of ionic PTMs, play a critical role in regulating protein interactions, localization, and activity. To test this hypothesis, our research objectives include:
Screening for PolyP-Modified Proteins
We will extend our screening efforts to identify additional histidine- and lysine-rich proteins that undergo polyP modification. Techniques such as NuPAGE and Western blotting will be employed to detect these modifications.
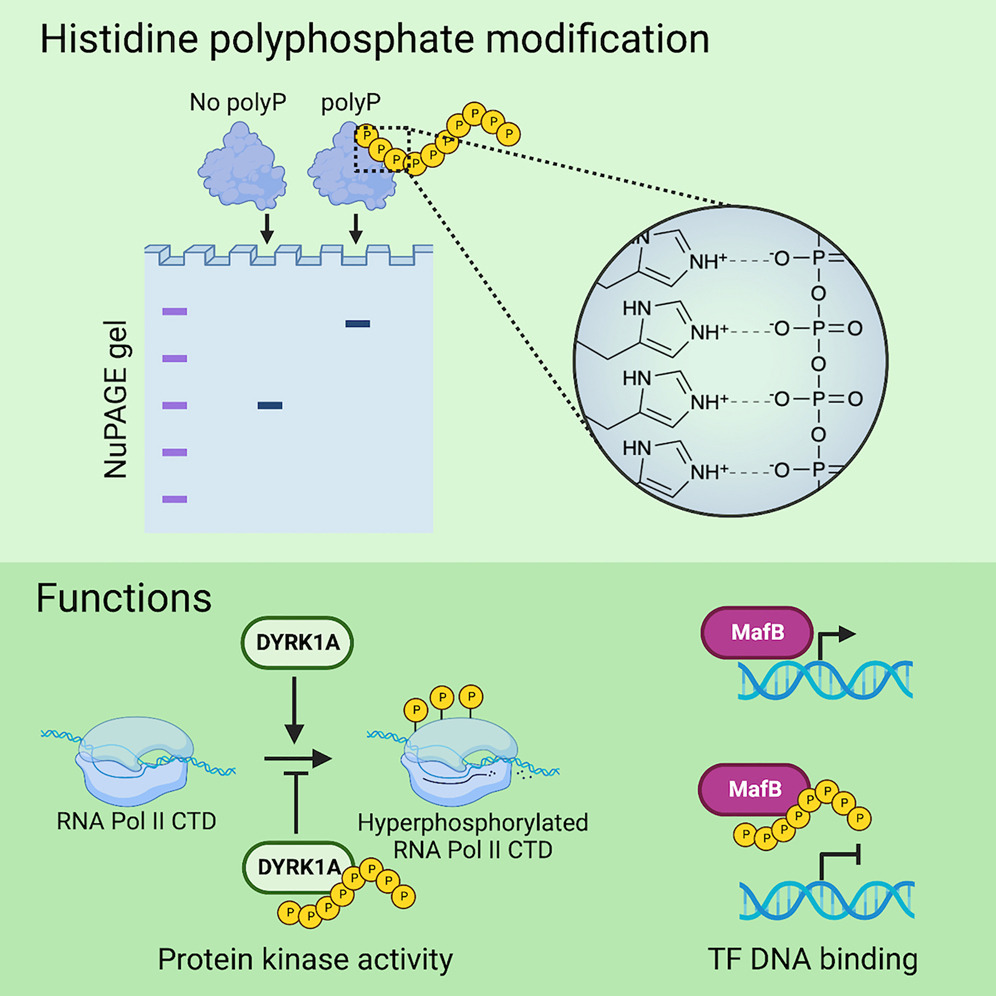
A striking role of polyP is its attachment to lysine residues via a non-enzymatic post-translational modification (PTM), previously believed to be covalent although likely not the case anymore.
Proteins with consecutive histidine residues undergo a similar modification by polyP, resulting in an electrophoretic mobility shift on NuPAGE gels.
Our screen identifies several dozens human and yeast histidine repeat proteins that experience histidine polyphosphate modification (HPM).
HPM disrupts phase separation and impairs the phosphorylation activity of the human protein kinase DYRK1A, as well as inhibits the function of the transcription factor MafB, suggesting HPM as a novel regulatory mechanism in proteins.
Characterizing PolyP-Protein Interactions:
We will utilize a range of biophysical and analytical techniques to further explore the non-covalent nature of PPM. This will include determining the binding affinities of polyP to histidine and lysine repeats, providing insights into the specificity and strength of these interactions.
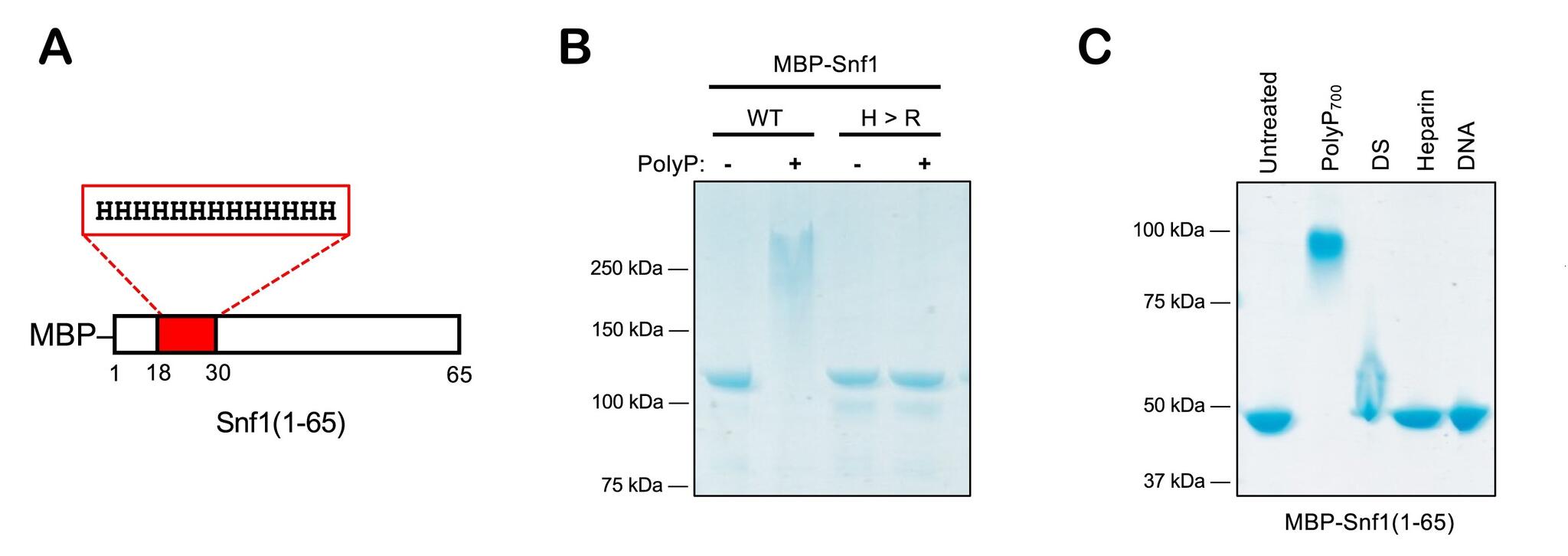
- Schematic of the MBP-Snf1(1–65) construct.
- Arginine replacement NuPAGE test.
- Polyanion NuPAGE test. DS, dextran sulfate.
Investigating Functional Implications
The functional consequences of polyP modifications will be studied in the context of transcription factors, kinases, and other regulatory proteins. Methods such as EMSA, RT-PCR, and Western blotting will be used to assess how these modifications impact protein function.
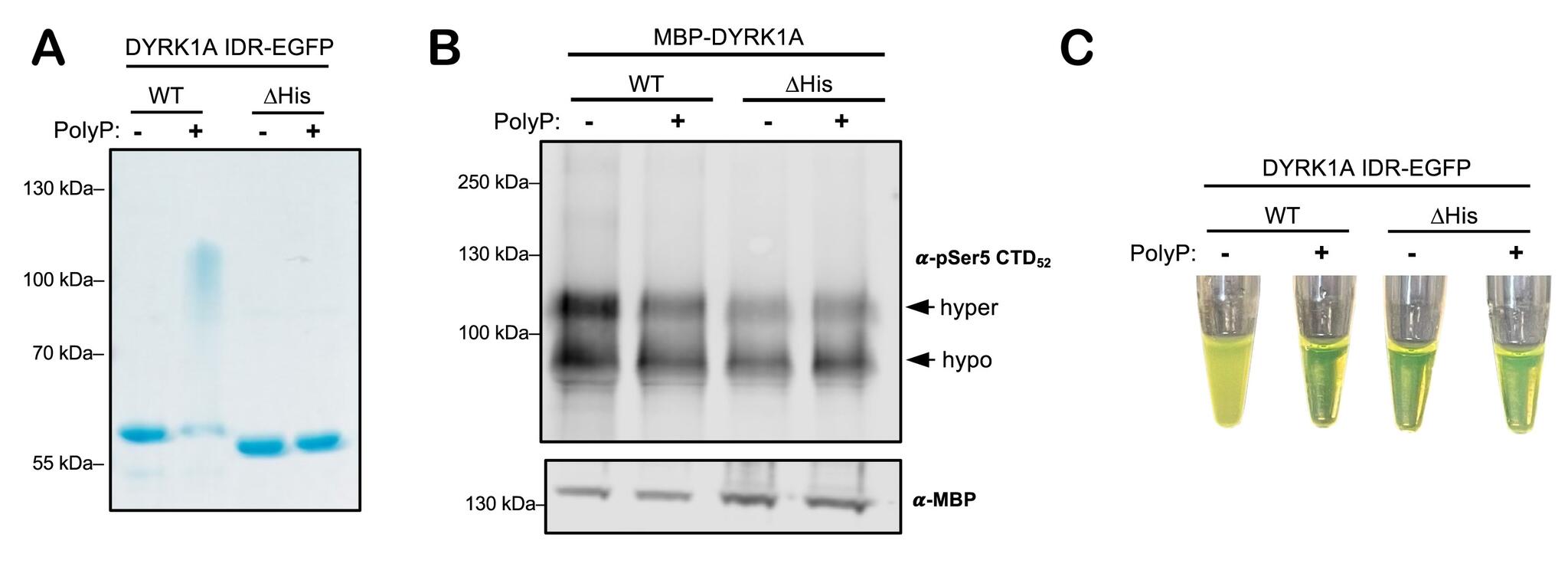
- NuPAGE analysis of samples.
- Kinase activity of MBP-DYRK1A on glutathione-S-transferase-CTD substrate detected via western blotting against phosphorylated CTD.
- Phase separation of WT or ΔHis DYRK1A IDR-EGFP pre-treated with polyP.
Examining Cellular Implications of PPM
Through cell biology experiments in both human and yeast models, we aim to uncover the broader biological significance of polyP modifications and their roles in cellular processes.
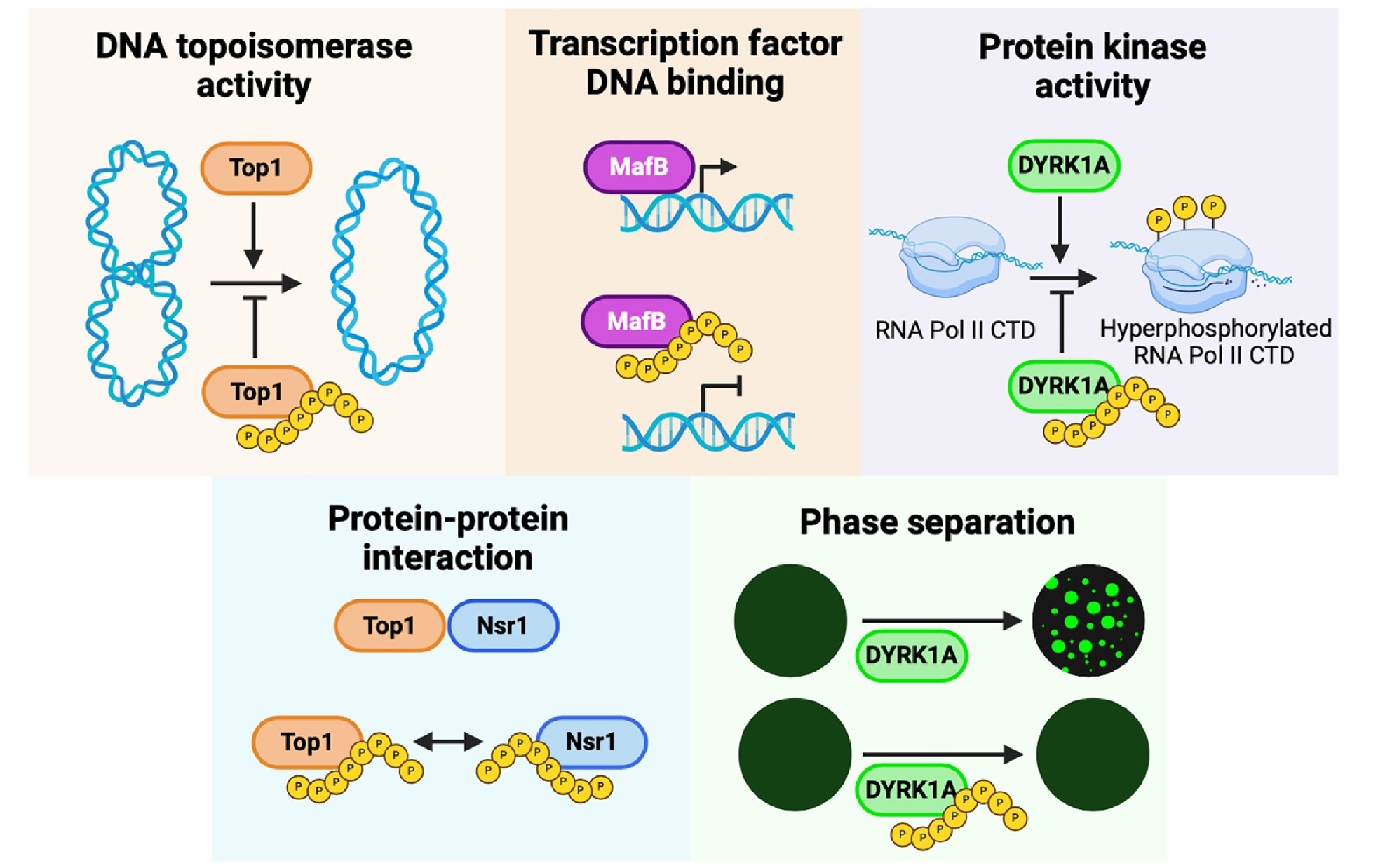
Schematic of lysine and histidine polyphosphate (polyP) modification and its effects on protein function.
Exploring Atypical Kinases
Our research will also delve into the regulation and mechanisms of atypical kinases, including alpha kinases. Leveraging AI-based approaches including de novo protein design, we will conduct an in-depth characterization of these kinases.
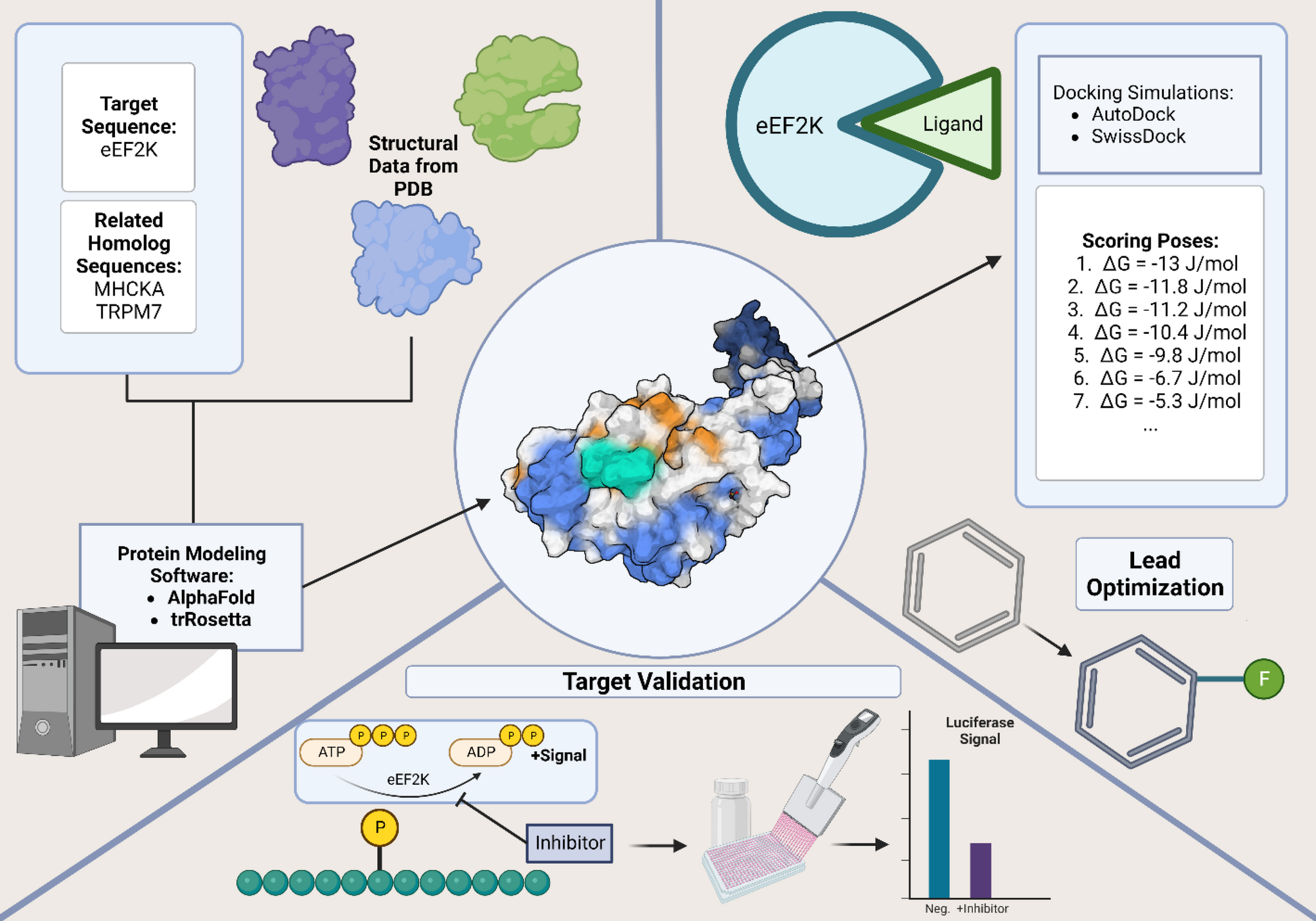
Simplified eEF2K inhibitor structure-based drug design pathway. The structure-based drug design process includes protein structure determination or accurate protein model development.
Protein-ligand interactions may be screened with docking simulations and leads are optimized.
In the case of eEF2K, a simple target validation may be executed to monitor the conversion of ATP to ADP and Pi using a luciferase assay.
Our research seeks to provide a comprehensive understanding of the role of polyP in protein function and regulation, offering new insights into the complexity of PTMs.