Our Research 'Philosophy' = "Metabolic Biochemistry Helps to 'Close the Gap' in Functional Genomics"
A thorough understanding of tissue-specific plant metabolism and its control is highly relevant to the optimal application of biotechnology for improving crops via metabolic engineering. Remarkable insights into plant metabolism are being provided by the ever-growing collections of plant genes and bioinformatic databases, as well as the implementation of high-throughput transcriptomic and mutant/transgenic studies.
While genomics provides a crucial blueprint and many powerful tools for systematic studies of metabolism, it simultaneously reveals that plant metabolism and its control is incredibly complex and poorly understood. In particular: (1) genome sequencing projects have revealed that a significant proportion of genes encode proteins having unknown functions, whereas many annotated genes encode multiple isozymes with poorly defined individual properties and roles, and (2) it is impossible to predict the functional properties of a protein or the kinetic and regulatory properties of an ENZ solely from genetic information.
With many genomes sequenced and others nearing completion, the next step is the less straightforward task of analyzing the expression and function of gene products (proteins, the vast majority of which = the ENZ catalysts), as well as more thoroughly elucidating metabolism and its control. The task of completing the picture of all cellular proteins, their actions and reactions, is one of the biggest challenges facing life science researchers today! Thus, genomics should be well integrated with parallel studies of the corresponding proteome, metabolome, in vivo metabolic fluxes, and membrane transporters/metabolic compartmentation, together with painstaking biochemical characterization of individual native ENZs.
Although under-represented in current plant science research, the native ENZ biochemistry research that is a foundation of the Plaxton lab helps to establish: 1) gene function, 2) enzyme physical, immunological, and kinetic/regulatory properties, 3) enzyme:enzyme interactions that may prevail in vivo, 4) enzyme transit/signal peptides and targeting, and 5) pivotal post-translational enzyme modifications such as phosphorylation, ubiquitination, or glycosylation.
The Functional Organization & Control of Plant Glycolysis
Glycolysis is the central metabolic pathway that fuels mitochondrial respiration, while producing ATP, reducing power, and biosynthetic precursors. Plant glycolysis is unique and complex as: i) it occurs in the cytosol and plastid with parallel reactions catalyzed by distinct isozymes, ii) cytosolic glycolysis is an intricate network of alternative enzyme reactions, & iii) its regulation is generally from the 'bottom up' with primary control at the level of phosphoenolpyruvate (PEP) metabolism. Thus, a long-time research emphasis has been on understanding the expression patterns, functions, localization, and molecular and biochemical properties of key glycolytic control enzymes, particularly those involved in PEP metabolism. Much of this research has been focused on the functions, and molecular and regulatory properties of unusual PEP carboxylase isoforms in developing vs. germinating castor oil seeds. Owing to its location at a crucial branchpoint in central plant metabolism, PEP carboxylase is tightly regulated by a diverse array of post-translational controls including protein:protein interactions, allosteric effectors, reversible phosphorylation, and monoubiquitination. This enzyme has become an attractive target for metabolic engineering of transgenic seeds having modified levels of storage oil versus storage proteins.

Model illustrating the biochemical complexity of developing vs. germinated castor oil seed (COS) PEP carboxylase (PEPC).
In developing castor beans the plant-type PEPC RcPPC3 exists as: (1) a typical Class-1 PEPC homotetramer which is activated in vivo by sucrose dependent phosphorylation of 50% of its p107 subunits (at Serine-11), & (2) part of the novel allosterically-desensitized Class-2 PEPC hetero-octameric complex with the multi-site phosphorylated bacterial-type PEPC RcPPC4 (p118) subunits. COS germination is accompanied by increases in RcPpc3 gene expression, PEPC activity and amount, and monoubiquitination of 50% of p107 subunits at Lysine-628 to form the novel Class-1 PEPC p110:p107 heterotetramer.
Metabolic Adaptations of Phosphate Starved Plants

We have studied the metabolic adaptations of plant respiratory metabolism to nutritional phosphate (Pi) starvation. This has enhanced our understanding of how the distinctive flexibility of plant cytosolic glycolysis and bioenergetics helps plants to acclimate to unavoidable environmental stresses such as anoxia and Pi starvation, & the unique role of pyrophosphate as an alternate cytosolic energy 'transmitter' to ATP. The identification and characterization of purple acid phosphatases (PAPs) upregulated by Pi-starved plants has also been of significant interest to us since secreted PAPs function to scavenge Pi from soil localized organic P-monoesters, whereas intracellular (vacuolar) PAPs remobilize Pi from intracellular P-monoesters and anhydrides.Our recent research in this area has included the complete purification and detailed biochemical and functional genomic characterization of the predominant Pi starvation upregulated intra- and extracellular PAPs of the model plant Arabidopsis thaliana. We have also been using a proteomics approach to discover novel Pi starvation inducible and differentially phosphorylated proteins of Pi-deprived Arabidopsis.
Model illustrating alternative pathways of cytosolic glycolysis, miETC, and tonoplast H+- pumping processes (indicated by bold arrows) that may facilitate respiration and vacuolar pH maintenance by Pi-deprived plant cells.
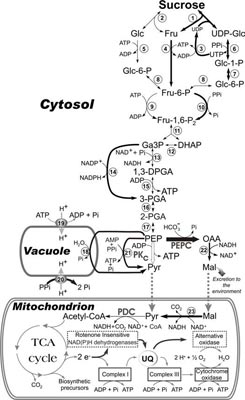
These bypasses negate any dependence of respiration and tonoplast H+-pumping on adenylates and/or Pi, the levels of which become markedly depressed during severe Pi starvation. Organic acids produced by PEP carboxylase may also be excreted by roots to increase the availability of mineral bound Pi. A key component of this model is the critical secondary role played by ‘metabolic Pi recycling systems’ during Pi deprivation.
Phosphite Blocks Plant Phosphate Sensing & Kills Phosphate-Starved Plants
 The Myth Of Phosphite Fertilizers
The Myth Of Phosphite Fertilizers
Over the past 30 years, a reduced form of Pi known as phosphite (Phi) has been widely used to improve the yield of many crops. Phi’s use in agriculture is highly controversial since it is being marketed both as a crop fungicide and as a superior source of crop phosphorus nutrition (relative to Pi). In the mid-1990s we began to assess the influence of Phi on plant growth and metabolism.
We discovered that low Phi concentrations are highly phytotoxic to Pi-starved, but not Pi-sufficient plants (and yeast) because Phi blocks the upregulation of Pi-starvation inducible enzymes and Pi transporters needed for plant acclimation to suboptimal Pi nutrition. Several top labs in the field of plant P-nutrition (including K.G. Raghothama at Univ. of Purdue, Steffen Abel at U.C. Davis, Deb Allan & Carroll Vance at Univ. of Minnesota ) have since employed Phi as a tool to help dissect plant Pi sensing pathways. The data in plants and yeast are consistent with the hypothesis that cell sensors and/or signaling components involved in the coordinated response to Pi-stress actually perceive Phi as Pi even though Phi is not metabolized. Nevertheless, farmers throughout the world are applying Phi formulations marketed as a "super P fertilizer" rather than as a fungicide; e.g., Nutri-Phite, Phosgard, & Nutramix, etc.
Agrochemical companies selling 'phosphite fertilizer' products appear to be avoiding the substantial expense and time associated with registering an agricultural fungicide (by labeling their Phi products as a "P fertilizer"). As a result of our phosphite research, The Fertilizer Section of the Canadian Food Inspection Agency (CFIA) banned sales of so-called ‘phosphite fertilizer’ products in Canada.
Research Tools & Recent Advances
A foundation for our lab's research has been the efficient use of fast protein liquid chromatography (FPLC) for the high-yield purification of active native enzymes, followed by detailed examination of their structural, immunological, and kinetic/regulatory properties. Thanks to NSERC, we have acquired many of the latest tools for purifying and characterizing enzyme proteins including GE Healthcare's state-of-the-art AKTA FPLC system, as well as two Molecular Devices P.C.-controlled kinetic microplate readers (that facilitate 'high throughput' enzyme activity determinations & kinetic studies).
However, over the past 15-20 years numerous innovative strategies have been incorporated into our research, particularly: 1) protein mass spectrometry for enzyme peptide mass fingerprinting, peptide sequencing, and PTM (phosphorylation-, glycosylation-, and monoubiquitination)-site mapping, 2) other key proteomic tools such as 2-dimensional and phosphate-affinity polyacrylamide gel electrophoresis, and co-immunopurification, 3) anti-phosphorylation-site specific-antibody production for enzyme phosphorylation and protein kinase studies, 4) isolation and use of relevant cDNAs as probes with respective antibodies to assess isozyme expression changes, and for recombinant enzyme expression in E. coli, and and 5) use of Arabidopsis T-DNA insertional 'loss of function' mutants to test the physiological/metabolic function(s) of several enzymes that we have thoroughly characterized at the biochemical level.
Thanks to the dedicated efforts of my recent students and post-docs, our collaborators, and our generous NSERC and Queen's Research Chair support these innovations have led to several remarkable discoveries that have set the stage for our current research. We have also integrated modern imaging equipment available in the Dept. of Biology at Queen's (including TEM and our Confocal Laser Scanning Microscope), with specific antibodies and molecular tools that our lab has generated to assess the in vivo subcellular location and protein:protein interactions of 'key' enzymes we have been studying.
_______________________________________________________________________________________________________________________________________
Collaborators (2000-Present)
Jay Thelen, Dept. of Biochemistry, Univ. of Missouri
Gustavo MacIntosh, Dept.of Biochemistry & Biophysics, Iowa State Univ.
Alisdair Fernie, Max Planck Instit. of Molecular Plant Physiology (Potsdam, Germany)
Glen Uhrig,Dept. of Biological Sciences, Univ. of Alberta (Edmonton), https://www.uhriglab.com/
Robert Mullen, Dept. of Molecular & Cellular Biology, Univ. of Guelph (Canada)
Wayne Snedden, Dept. of Biology, Queen's University (Canada)
Jacqueline Monaghan, Dept. of Biology, Queen's University (Canada)
John Lunn, Max Planck Institute of Molecular Plant Physiology (Potsdam, Germany)
Shelia Macfie, Dept. of Biology, Western Univ., https://www.uwo.ca/biology/faculty/macfie/
Maria Fernandez, Dept. of Physical, Chemical and Natural Systems, Universidad Pablo de Olavide (Sevilla, Spain)
Pascale Champagne, Dept. of Civil Engineering, Queen's University
Mike Shane, School of Plant Biology, Univ. of Western Australia
Yi-Min She, Shanghai Instit. for Plant Stress Biology, Chinese Academy of Sciences
Cristina Echevarria, Dept. de Biologia Vegetal, Universidad de Sevilla (Spain)
Thomas McKnight, Dept. of Biology, Texas A & M Univ.
Daowen Wang, Institute of Genetics and Developmental Biology, Chinese Academy of Sciences, Beijing (China)
Norm Huner, Dept. of Biology, Univ of Western (Canada)
Craig Leach, Progenra Inc., Malvern, PA (USA)
Florencio Podestá, CEFOBI, Univ. of Rosario (Argentina)
Zongchao Jia, Dept. of Biochemistry, Queen's University (Canada)
Susan Wood, Dept. of Biology, Queen's University (Canada)
David Turpin, University of Alberta (Canada)
Chaofu Lu, Montana State University (USA)
Alex Valentine, Dept of Biotechnology, Univ. of the Western Cape (South Africa)
Justin MacDonald, University of Calgary (Canada)
Bruce Grant, Dept. of Biochemistry, Univ. of Melbourne (Australia)
Julie Niere, Royal Melbourne Instit. of Technology(Australia)
Eduardo Blumwald, UC Davis (USA)
KG Raghothama, Purdue University (USA)
Arvind Kayastha, Banaras Hindu University (India)