Bio-organic and bio-inorganic projects: Targeting DNA and RNA
The genetic information contained in our DNA defines what we look like and how our body functions. It resides in several features:
- the atomic information : expressed through the hydrogen bonding pattern on each nucleic base; this is included in the primary structure,
- the local information : expressed in the conformation adopted by a fragment of DNA; this defines secondary structures such as the A, B and Z duplexes, quadruplexes, four-way junctions…
- the deviations from ideal secondary structures, which may include stacked non-base-paired nucleic bases in a duplex, flipped-out bases, bending and untwisting.
- the overall packaging (e.g. chromosome compaction).
All these features are a source of information for the recognition of DNA.
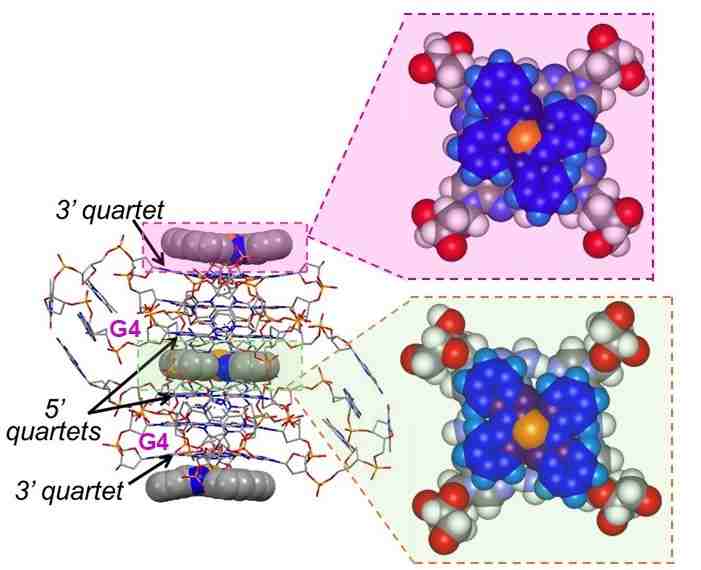
In the lab we design, synthesize and study organic and inorganic DNA targeting sensors based on each of the first three features. These indeed mark important sites of either “healthy DNA” that we want to target (e.g., telomeres) or damaged DNA. In particular we are interested in detecting molecular and supramolecular signatures related to cancers. In the latter case, the goal is to use our targeting agents to yield a signal that may be detected with a physical response (e.g. luminescence for diagnostics), or by proteins and enzymes for in situ biological responses. Selective targeting of special DNA secondary structures that are different from traditional B-helix DNA, here guanine quadruplexes, is illustrated below.
In addition to designing binders, we are also interested in letting the target DNA select its own preferred substrate from a library of dynamically exchanging potential binders (why should we do all the work ?!).
Dynamic chemistry: Molecular and supramolecular projects:
-
Self-assembled responsive artificial ion channels: self-assembly is a process that lets a molecule assemble according to the electronic information within its structure, as is typically illustrated by DNA A, B and Z duplexes. As the interactions are dependent on the environment (solvent, salt concentration…), so is the outcome of the assembly. We are using this stimulus-dependent organization of matter to assemble artificial ion channels whose structure should depend on voltage, light, solvent, or salt concentration. The design of our molecular building blocks also allows fixing the assembly into easily characterizable covalent architectures.
-
As is illustrated by DNA self-assembly, the hierarchy of information involved in the formation of complex architectures is of interest (hydrogen bonding, then pi-stacking and salt effect…). Hence, small projects in the lab focus on defining pathways for a cascade of interactions to take place and yield highly complex architectures from very simple building blocks. Sometimes, the self-assembled products exceed expectations !
